 Protein Knots
Protein Knots
 Protein Knots
Protein Knots
List of known knots
| Protein | Species | PDB code | Length | Knot | Knotted core |
|---|---|---|---|---|---|
| YbeA-like | E.coli | 1ns5 | 153 | 31 | 69-121 (32) |
| T.maritima | 1o6d | 147 | 31 | 68-117 (30) | |
| S.aureus | 1vh0 | 157 | 31 | 73-126 (31) | |
| B.subtilis | 1to0 | 148 | 31 | 73-125 (32) | |
| tRNA(m1G37)-methyltransferase TrmD | H.influenza | 1uaj | 241 | 31 | 85-130 (92) |
| E.coli | 1p9p | 235 | 31 | 90-130 (89) | |
| SpoU-like RNA 2'-O ribose mtf. | T.thermophilus | 1v2x | 191 | 31 | 96-140 (51) |
| H.influenza | 1j85 | 156 | 31 | 77-114 (42) | |
| T.thermophilus | 1ipa | 258 | 31 | 190-234 (29) | |
| E.coli | 1gz0 | 242 | 31 | 173-215 (28) | |
| A. aeolicus | 1zjr | 197 | 31 | 100-144 (58) | |
| S. viridochromog. | 1x7p | 267 | 31 | 209-251 (31) | |
| YggJ C-terminal domain-like | H.influenza | 1nxz | 246 | 31 | 166-217 (30) |
| B.subtilis | 1vhk | 235 | 31 | 168-226 (27) 1 | |
| T.thermophilus | 1v6z | 227 | 31 | 104-203 (25) 3 | |
| Hypothetical protein MTH1 (MT0001) | A.M. thermoautotr. | 1k3r | 262 | 31 | 48-234 (28) |
| Carbonic Anhydrases: | |||||
| Carbonic Anhydrase | N. gonorrhoeae | 1kop | 223 | 31 | 39-226 (0) |
| Carbonic Anhydrase I | H.sapiens | 1hcb | 258 | 31 | 29-256 (2) |
| Carbonic Anhydrase II | H.sapiens | 1lug | 259 | 31 | 31-257 (3) |
| Bos Taurus | 1v9e | 259 | 31 | 32-256 (3) | |
| Dunaliella salina | 1y7w | 274 | 31 | 39-272 (4) | |
| Carbonic Anhydrase III | Rattus norv. | 1flj | 259 | 31 | 31-258 (3) |
| H. sapiens | 1z93 | 263 | 31 | 31-258 (9) | |
| Carbonic Anhydrase IV | H.sapiens | 1znc | 262 | 31 | 31-258 (1) |
| Carbonic Anhydrase V | Mus musculus | 1keq | 238 | 31 | 31-258 (4) |
| Carbonic Anhydrase VII | H.sapiens | 1jd0 | 260 | 31 | 31-258 (3) |
| Carbonic Anhydrase XIV | Mus Musculus | 1rj6 | 259 | 31 | 31-258 (2) |
| Miscellaneous: | |||||
| Ubiquitin Hydrolase UCH-L3 | H.sapiens | 1xd3 | 229 | 52 | 13-212 (12) 4 |
| S.cerevisiae (synth.) | 1cmx | 214 | 31 | 14-228 (6) 4,1 | |
| Ubiquitin Hydrolase UCH-L1 | H.sapiens | 2etl | 219 | 52 | 10-216 (13) |
| S-adenosylmethionine synthetase | E.coli | 1fug | 383 | 31 | 33-260 (32) |
| rattus norv. | 1qm4 | 368 | 31 | 46-281 (29) 1 | |
| H. sapiens | 2hj2 | 380 | 31 | 37-280 (21) | |
| Class II ketol-acid reductoisomerase | Spinacia oleracea | 1yve | 513 | 41 | 321-533 (62) |
| E.coli | 1yrl | 487 | 41 | 222-437 (52) 2 | |
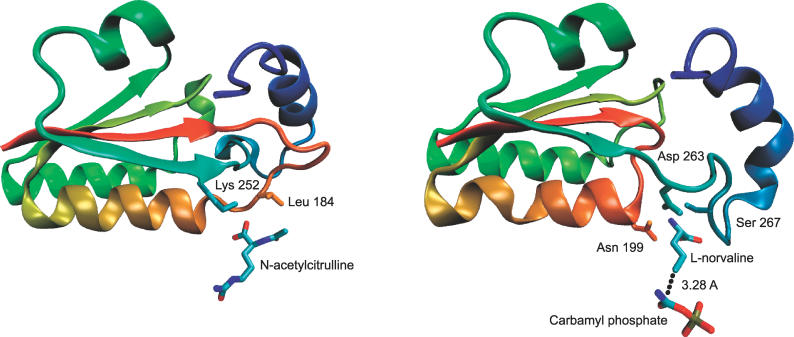
| Transcarbamylase | B.fragilis | 1js1 | 324 | 31 | 169-267 (57) |
| X.campestris | 1yh0 | 328 | 31 | 173-277 (57) 2 | |
| Methyltransferase | H.sapiens | 2ha8 | 159 | 31 | 103-148 (30) |
| P. gingivalis | 2i6d | 231 | 31 | 177-223 (9) |
How we define knots
Mathematically, knots are only well defined in closed loops. However, the ends of proteins are usually located on the surface and can be connected unambiguously. A thus connected loop can be tested for knots with the Alexander polynomial.
In the course of our investigations we came up with a number of stringent
criteria which a knotted structure should fulfill:
Unfortunately, there are many incomplete structures with gaps. If the structure is incomplete a knot should be present in at least one fragment and the structure which results when gaps are bridged with straight lines.
Questions about particular knots? Contact Peter Virnau.
![]() Examples
of different knot types:
Examples
of different knot types:

![]() Evolutionary
origin:
Evolutionary
origin:
Structures of Transcarbamylase from X. campestris with a Trefoil Knot and from Human without a Knot