Results & Experiments
![]() Specificity of TCRs and transgenic mice:
Specificity of TCRs and transgenic mice: 
E.S. Huseby, F. Crawford, J. White, P Marrack & J.W. Kappler, Nat Immunol 7, 1191 (2006)
Created transgenic mice, expressing only one self-peptide (M=1) in their thymus!
Compared specificity of TCRs from normal and transgenic mice to mutantations of a recognized peptide:
Normal mice TCR typically do not recognize peptides after single site mutations,
while transgenic mice TCR are tolerant of most single site mutations, except at a few "hot spots".
To model this, simulations were performed for TCRs of length N=5 and varying values of M
(Typical number of peptide encounters in thymus is 1,000-10,000)
In normal mice, negative selection is dominant, increasing the frequency of weak amino-acids in mature TCRs, while
in transgenic mice, positive selection enriches strong amino-acids.
In normal mice TCR/peptide binding is due to many weak bonds, single site mutations can abrogate recognition,
in transgenic mice TCR a few strong bonds suffice for recognition, other sites are mutationally tolerant.
![]() Elite controllers of HIV appear to better tolerate mutations of the virus. Is a similar mechanism at play?
Elite controllers of HIV appear to better tolerate mutations of the virus. Is a similar mechanism at play?
A. Košmrlj, ..., A.K. Chakraborty, Nature online (5 May 2010) WebMed, ...
![]() Enrichment of TCRs in weak amino-acids?
Enrichment of TCRs in weak amino-acids?
In 2008, we examined a limited set ot then available crystallographic data from TCR/peptide interfaces:
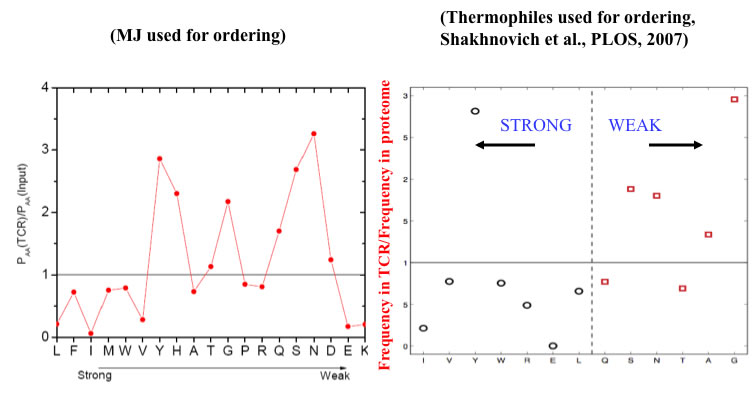
A recent analysis of a large database of TCR sequences did not reveal such bias, but did not distinguish peptide contacts.
Y. Elhanati, A. Murugan, C.G. Callan,T. Mora, & A.M. Walczak, PNAS 111, 9875 (2013) (offlline)
![]() Self-reactive TCR (implicated in autoimmune disease) are enriched in stronger binding amino-acids:
Self-reactive TCR (implicated in autoimmune disease) are enriched in stronger binding amino-acids:
B.D. Stadinski, K. Shekhar, ..., A.K. Chakraborty & E. Huseby, Nature Immunology 17, 946 (2016)
Interpreting and accounting for nonuniform TCR contact profiles
Hanrong Chen, A.K. Chakraborty, & M. Kardar, (2018) (offlline)
(1)
vs. (2)
While the mean energy is the same in the two cases, the variance in energy is different.
The (experimentally seen) enrichment of weak amino-acids at sites 6 & 7, is observed in model (2) but not (1).