Massachusetts Institute of Technology
6.891 Computational Evolutionary Biology
Laboratory 2: From molecular to genomic evolution
Handed out: November 15 Due: December 1
Introduction
In this laboratory we shall apply evolutionary models to genomic data – from the very lowest levels, nucleotides and amino acids, to the question of whole-genome duplication. Our major goal will be to have you become familiar with modern methods for detecting natural selection at the level of genes and proteins, as well as to engage some current recent questions in gene duplication. The lab will consist of four parts: the first three will have you get familiar with the tools, using small or simulated data-sets, while the fourth part will turn to the yeast data set studied in Kellis et al. (2003, 2004). You should complete the first three parts before turning to the fourth. (There is a lot to do in this laboratory!)
Part 1: Warmup exercise with coalescent processes
The purpose of this exercise is
to illustrate the coalescent process with various extensions in the discrete as
well as the continuous form. We will use two small java-applets that can
“animate” the coalescent process, the Wright-Fisher animator (discrete
generations) and the Hudson-animator (continuous time). These tools can both be accessed
through http://www.coalescent.dk.
It is important that you take
time to think about the different questions while doing each exercise –
these are designed to help you build intuition about the coalescent process.
You might well ask what
coalescent models are used for. That’s a great question. As one example, they are used to get
confidence intervals in situations where we cannot compute exact statistical
distributions. This is done in the
DnaSP program, for instance. The
coalescent is used to simulate a large variety of possible genealogical
outcomes.
Part 1.1. The Wright-Fisher
animator
By following the link
Wright-Fisher animator at www.coalescent.dk
it is possible to use the Wright-Fisher animator to follow the reproduction
process forward in time and the coalescent process backwards in time one
generation at a time. After you have followed the reproduction process a number
of generations forwards in time it is possible to “untangle” the genealogy, and
then to follow both how many descendants each of the original genes leave over
generations (click on upper row), and to follow the ancestors to the sequence
in the bottom row (by pressing the circles in the bottom row).
A new simulation is carried out
by setting the parameters for the number of generations, g and the number of genes, n, and then and pressing the “New” button. The
simulation can then be controlled via the buttons in the bottom (right) part of
the window, e.g. one generation at a time. One button enables you to untangle
the resulting genealogy (i.e. rearranging individuals so that lines do not
cross). The ‘fastforward’ skips ahead
and does the whole simulation (all generations at once). A green
1. Set n=number of genes(= ‘individuals’ here) =10, and g=number of generations=15.
a. Forward
in time: How many generations does each of the initial genes persist in the
population?
b. What
is the expected number of generations for persistence?
c. Backwards
in time (coalescence). Do all the genes coalesce within this time span? If so, what is the number of
generations to coalescence of the sample? If not, how many ancestors are there
and how large part of the sample are they ancestors to?
d. What
would you have expected from theory?
2. Repeat
this exercise 5 times, writing down the number of ancestors and the time to
coalescence. This should give you an idea about the variance of the process.
How is the variance in time going from 3 to 2 ancestors related to that going
from 2 to 1 ancestor? How does
this compare to theory?
3. Try
the first exercise with different values of n and g. How does the time
to coalescence scale with n? Again compare to theory.
Part 1.2. The Hudson
animator.
The Hudson-animator is reached
through www.coalescent.dk,
and then choosing “Hudson animator.” This is the continuous-time version
of the discrete ‘atom-based’ Fisher-Wright model. It can carry out the following simulations:
Š The
basic coalescent and the Coalescent with recombination
Š Coalescent
with exponential population growth
Š Coalescent
with migration
Š Coalescent
with directional selection (the ancestral selection graph)
Please consult the manual
before doing the exercises, it can be found under help at the start page. It
briefly describes how to control the applet.
Coalescent with
recombination.
1. The
basic coalescent. The
Fisher-Wright model assumes a non-sexually reproducing organism. What happens if there is mixing via
genetic recombination? Select the “recombination” link after choosing the
Hudson animator, or make sure the ‘recombination’ pull-down menu item is
selected. Choose n=5 sequences and rho=0 (no
recombination). Press recalc and the animation starts. The speed can be
controlled via the “speed” item on the bottom-left corner, or you can just
fast-forward as before. Details regarding each node, e.g., the scaled time to
coalescence, can be viewed in the small window at the right when moving the
mouse over the relevant node.
Clicking on the ‘trees’ panel will untangle the trees, as before.
a. What
is the time to the first coalescence event? Write it down.
b. What
is the time to the most recent common ancestor? Write it down.
c. Repeat
(a) and (b) 5 times (by pressing recalc). How does the time to first coalescence
and time to most recent common ancestor vary?
2. Repeat
with n= 10 and n=20 sequences. What are the times to the first
coalescence and the most recent common ancestor in these cases? Write this down
for 5 different runs.
The recombination rate is
determined by rho=2nc, where c is
a constant. In the animation,
recombination events are marked as blue nodes (in contrast to the green nodes
of coalescent events). Now we’ll
see what the mixing effect of recombination does to genealogy trees.
4. Set n=5,
and rho=1. Press recalc and study the animation. Do any recombination events
occur? These will be marked by
blue nodes, along with a crossing line to the other individual with whom the
recombination occurred. Also, if you mouse over the blue node, you’ll see what
‘fraction’ of that individual’s ‘genetic’ material that was exchanged (see
picture below). If not, try again.
Try a total of 5 times and write down the number of recombination events
in each case. Examine where in
the sequence recombination events occur.
a. Are
there any examples where a different part of the sequence of ‘individuals’ has different most recent common ancestors (MRCAs, marked in
green) at different time
points? Why would this be possible?
b. Can
you find any examples of “trapped” non-ancestral material (colored white)
between blocks of ancestral material?
c. Try
to press the ‘trees’ tab. Here it is possible to study each of the different
trees over the sequence. How many different trees are there and how does this
relate to the number of recombination events? Try to find examples of each of
the following recombination types:
i. Recombination
changing the topology of the tree
ii. Recombination
changing the branch length but not the topology of the tree
iii. Recombination
that does not change the tree
What is the total number of
each of types i, ii, and iii during the 5 replicate runs?
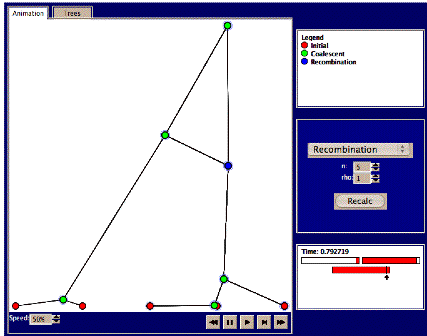
5. Try
setting rho=2 and n=10
a. How
many recombinations occur now?
b. At
which time do different parts of the sequence find a most recent common
ancestor?
c. At
which time have all the different parts of the sequence found a most recent
common ancestor? Compare this time with the time to most recent common ancestor
without recombination.
5. Try
setting rho=5 and n=20. How many
recombination events occur now? How many different most recent common
ancestors are there over the sequence now?
Discuss in a few sentences the
implications
Coalescent with exponential
growth
Now choose the coalescent
animation with exponential growth selected on the pull-down menu. This is
controlled with the parameter exp, which
is equal to Nb. This parameter
measures how many times the present population is larger than the population 2N, where N=
the size of the present day population. For humans, all estimates suggest that exp>100.
6. Try
to simulate n=10 sequences and different
runs with exp=1, 10, 100, 1000
a. How
does the shape of the genealogical tree depend on the value of exp?
b. How
can that be?
Part 2: Detecting selection – simple methods
Part 2.1 Detection of selection in primate lysozyme
enzymes
Already-aligned lysozyme C precursor DNA sequences from
different species of apes (including human) in FASTA format may be found here. Analyze the pattern
of nonsynonymous and synonymous substitutions between these genes. Use the SNAP
utility at the HIV database, http://hiv-web.lanl.gov/SNAP/WEBSNAP/SNAP.html.
1.
Using the nonsynonymous/synonymous ratio test, is there any
evidence for positive selection on the lyz C gene in ape-like primates?
2.
What is the overall dN/dS ratio for the lyz C genes in the
alignment? Is there an excess of silent or coding mutations?
3.
Are the phylogenetic trees based on dN and dS different? What do you think this means?
4.
Discuss the results of this analysis. Why might the lyz C
genes among these species differ? Can natural selection be responsible? (Reference: Polley SD, Conway DJ. 2001.
Strong diversifying selection on domains of the Plasmodium falciparum apical
membrane antigen 1 gene. Genetics.
158:1505-1512.)
Part 2.2 Detection of selection with Tajimas's D test
Fetch this sequence alignment of Plasmodium falciparum (malaria) apical membrane antigen 1 gene sampled
according to several sources (here) and save the
sequences on your local computer.
(For the next part we assume that you all have access to PCs running
Windows…if not, see me.) Download
and install DnaSP on your local
PC; the web site is http://www.ub.es/dnasp/dnasp406.exe (The main web site is http://www.ub.es/dnasp/) This file is a self-extracting, and you
just run it to install the program itself. You will use the program to analyze nucleotide diversity and
test selection with respect to this dataset via “Tajima’s D” using a sliding window of appropriate size.
“Tajima’s D” tests the statistical significance of
an excess of common as compared to rare
nucleotide variants, via the difference between two different estimates of
nucleotide diversity. Under the neutral model, i.e., constant population size
and no selection, we can set one estimate as θ= 4Nμ. We can
make a second estimate of nucleotide diversity as Ļ= the sum of the number of
nucleotide differences between the ith
and the jth DNA sequences. D is simply Ļ-θ, and the null hypothesis is that
this value is 0. Departures from 0
indicate that the underlying assumptions might be incorrect – i.e., a
negative value of D indicates
either purifying selection or population expansion, while positive values of D indicate positive selection.
You can use DnaSP as follows:
Š Start
the program DnaSP and open the faliciparum.fas
file. The program will put up a
short summary window, which you can then close
Š Select
“Overview” and run the program to obtain summary statistics that include an
estimated number of ‘haplotypes’
as well as summary calculations of selection neutrality (Tajima’s D, Fu and
Li’s D*, etc.) – record these. Note that these are calculations for the
gene as a whole.
Š Carry
out a sliding window analysis of the nucleotide diversity: under “Analysis”
select “Tajima’s Test” and then from resulting pop-up Options window, look on
the right where one selects a ‘sliding window’ analysis, so that one can see
how D varies from gene region to gene
region. Check the ‘Compute’ box – but you will have to do some
experimenting to figure out the step size should be (try the default
first?). After you click “OK” you
will get a pane that has text output and a table. The table will have a
check-box to produce a graph that plots nucleotide (DNA) portion on the X-axis
and Tajima’s D on the
Y-axis. See if you can locate any
region in the gene that seems to be under selection.
Š What form of selection is this?
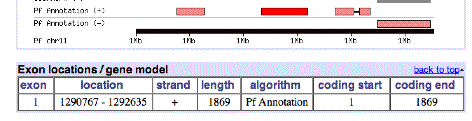
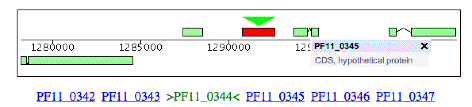
Here is some information on this gene, P. falciparum PF11_0344, an apical membrane precursor, 622 amino
acids long. It resides on
Chromosome 11 of P. falciparium,
spanning locations 1290767 – 1292635. I have placed a link to the NCBI entry on this gene
and a link to the plasmodium database entry for this gene below. You can use these links to view the
gene’s amino acid sequence, nucleotide sequence, and even the 3-D structure of
part of the corresponding protein (structure 1HN6). Some snapshots from the plasmodium database are below. Perhaps the best description is the
link that follows, from the Sanger Center, which gives a good overview of the
functioning of the protein associated with this gene and a great browser view.
Link 0: Sanger Center Plasmodium falciparum database
Link 1 to the NCBI genome database: http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&val=23508535
Link 2 to the Plasmodium database: http://plasmodb.org/plasmodb/servlet/sv?page=gene&source_id=PF11_0344
Link 3 to the Plasodium database genome browser:
http://plasmodb.org/cgi-bin/plasmodb/servlet/genomicSeqBrowser.pl?geneSourceId=PF11_0344&taxonId=211
Link 4 to the 3-D structure PDB database:
http://www.rcsb.org/pdb/cgi/explore.cgi?job=graphics&pdbId=1HN6&page=
Link 5 to the Conserved Domain Database (CDD) at NCBI:
http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?uid=2940
Chromosome 11 picture:
Gene AA sequence:
![]()
The amino acid sequence for this gene is as follows (from NCBI):
1 mrklycvlll safeftymin
fgrgqnyweh pyqnsdvyrp inehrehpke yeyplhqeht
61 yqqedsgede ntlqhaypid
hegaepapqe qnlfssieiv ersnymgnpw teymakydie
121 evhgsgirvd lgedaevagt qyrlpsgkcp vfgkgiiien snttfltpva tgnqylkdgg
181 fafppteplm spmtldemrh fykdnkyvkn ldeltlcsrh agnmipdndk nsnykypavy
241 ddkdkkchil yiaaqenngp rycnkdeskr nsmfcfrpak disfqnytyl sknvvdnwek
301 vcprknlqna kfglwvdgnc ediphvnefp aidlfecnkl vfelsasdqp kqyeqhltdy
361 ekikegfknk nasmiksafl ptgafkadry kshgkgynwg nyntetqkce ifnvkptcli
421 nnssyiatta lshpievenn fpcslykdei mkeiereskr iklndnddeg nkkiiaprif
481 isddkdslkc pcdpemvsns tcrffvckcv erraevtsnn evvvkeeykd eyadipehkp
541 tydkmkiiia ssaavavlat ilmvylykrk gnaekydkmd epqdygksns rndemldpea
601 sfwgeekras httpvlmekp yy
The full ORF sequence (so far) of the entire falciparum chromosome 11 may be found here in fasta format: http://web.mit.edu.ezproxyberklee.flo.org/6.891/www/lab/Pfa3D7_chr11ORFs50.fasta
(This includes just predicted ORFs larger than 50 amino acids long.)
The full nucleotide sequence for gene AMA1 (PF11_0344) may be found here:
http://web.mit.edu.ezproxyberklee.flo.org/6.891/www/lab/pf11_0344.fasta
Part 2.3. Volatility. Given this information, we can now test in a noncomparative way whether this gene is under positive selection: you can use J. Plotkin’s volatility server, at http://volatility.cgr.harvard.edu/cgi-bin/volatility.pl, and run it over all of the ORFs in chromosome 11 to see whether the PF11_0344 gene stands out. (The server takes fasta format as input, so you can use the chromosome 11sequence in the lab directory, above.) Please go ahead and do this now. Go to the volatility server and upload the entire chromosome 11 sequence. What should the kappa (transition/transversion rate ratio) be? (Click on the ‘About kappa’ box. See if you can figure out how to use DnaSP to estimate kappa, or do a bit of research on how this is done.) After you run the program, you can download the result to an excel spreadsheet locally. Then you’ll have to do a bit of grepping to find the AMA1 gene (but it’s not that hard). See what its computed volatility is – does it agree with your previous analysis from section 2.2? What do you think this means?
Part 2.4. Going further. Let’s do a bit more digging on this one – a more open-ended question. The analysis in section 2.2 looked at parts of a single gene, while the volatility analysis compares whole genes against the rest of a genome. See if you can use DnaSP to locate the regions within the gene that seem to be undergoing selection, and look at which amino acid residues they correspond to. Can you locate them in the actual 3-D structural map that the PDB provides? (I know you might have to ask your resident bioinformatics person in class about some of the details about how to do this, or the biochemistry – that is OK – just do your best and please work together.)
Part 3. Detecting selection – likelihood methods
(PAML)
Part 3.1
In this exercise, we shall use
likelihood methods to test the sensitivity of dN/dS, aka ‘omega’, with respect
to basic evolutionary assumptions about codon evolutionary changes.
Dataset: GstD1 genes of Drosophila
melanogaster and D. simulans (600 codons). This data file is located here.
Objective 1: Test the effect of transition / transversion ratio
(kappa, orκ) on dN/dS (Note
that you use this ratio when you employ J. Plotkin’s volatility analysis. Hmm - perhaps you can use this estimation in the previous
problem.)
Objective 2: Test the effect of codon evolutionary change
differences (ĻI’s).
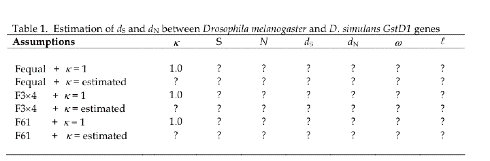
Your goal will be to fill in
the following table, which displays six rows, corresponding to six possible
combinations for kappa and pi:
All codon frequency changes equal
(denoted Fequal), and kappa = 1.
All codon frequency changes
equal, and kappa estimated by maximum likelihood from the actual data
Codon frequencies estimated via a
Jukes-Cantor 3x4 redundancy class matrix, and kappa = 1
Codon frequencies estimated via a
Jukes-Cantor 3x4 matrix, and kappa estimated from the data
Full 61x61 codon frequency
matrix, and kappa = 1
Full 61x61 codon frequency
matrix, and kappa estimated from the data
(See the Lewontin paper for further discussion of these
codon evolution matrices.)
Terminology:
κ=
transition/transversion rate ratio
S = number of
synonymous sites
N = number of
nonsynonymous sites
ω= dN/dS
ℓ= log
likelihood score (selection vs. no selection)
Your filled in table should be something like this, with
numbers replacing the question marks:
The general PAML web page may be found here:
http://web.mit.edu.ezproxyberklee.flo.org/6.891/www/lab/paml.html
You can read the documentation file (rather opaque), in pdf,
here: http://web.mit.edu.ezproxyberklee.flo.org/6.891/www/lab/pamlDOC.pdf
The actual PAML site has an FAQ that is worth reading if you
want to really delve into things, or seek more clarification.
The control data file for the PAML program (see web page here
– download and edit).
---------------------- beginning of file -------------------------
seqfile = seqfile.txt * (DNA) sequence data
filename (replace)
outfile = results.txt *
main result file name (replace w/ yours)
noisy = 9 *
0,1,2,3,9: how much output on screen
verbose = 1 *
1:detailed output
runmode = -2 *
-2:pairwise comparison
seqtype = 1 *
1:codons
CodonFreq = 0 *
0:equal,1:F1X4,2:F3X4,3:F61[CHANGE THIS]
model = 0 *
NSsites = 0 *
icode = 0 *
0:universal code
fix_kappa = 1 * 1:kappa fixed, 0:kappa to be
estimated * [CHANGE THIS]
kappa =
1 *
fixed or initial value
fix_omega
= 0 *
1:omega fixed, 0:omega to be estimated
omega = 0.5 *
1stfixed omega value
*
Uncomment out the various lines below to get the various combos
* Codon
bias = none; Ts/Tv bias = none
* Codon
bias = none; Ts/Tv bias = Yes (ML)
* Codon
bias = yes (F3x4); Ts/Tv bias = none
* Codon
bias = yes (F3x4); Ts/Tv bias = Yes (ML)
* Codon
bias = yes (F61); Ts/Tv bias = none
* Codon
bias = yes (F61); Ts/Tv bias = Yes (ML)
---------------
end of control file input -------------------------
Discuss your findings with respect to the sensitivity of the analysis to various assumptions about nucleotide changes – do the assumptions make a difference in this case? ‘
Part 2. Likelihood ratio test method for detecting
variation in seletion pressure among branches in a phylogenetic tree.
Example: The lactate dehydrogenase (Ldh)
gene family is an important model system for investigating the molecular
evolution of isozyme multigene families.
The rate of evolution for Ldh is
known to have increased in the form Ldh-C following a gene duplication event. (See reference: Lactate dehydrogenase (LDH) gene
duplication during chordate evolution: the cDNA sequence of the LDH of the
tunicate Styela plicata. Stock
DW, Quattro JM, Whitt GS, Powers DA., Mol Biol Evol.1997 Dec.14(12):1273-84).
You can take the following tree
as indicative of hypotheses we would like to investigate:
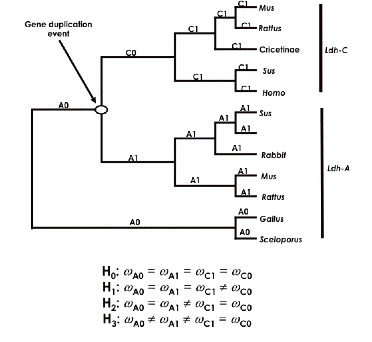
Here we hypothesize that the ancestral form duplicated into
two types: Ldh-A and Ldh-C. We want to use PAML to see if we can answer the
following questions:
1.
Was there an increase in the underlying mutation rate of Ldh-C?
2.
Has there been increased positive selection following the
duplication event?
3.
Has there been a long term change in selection pressure?
These are all questions regarding
the relative differences among selection
between different branches of this presumed phylogenetics tree. Using PAML, we
want to compare the relative selection pressure on these two branches via a
likelihood ratio test – what PAML calls a ‘branch specific’ codon model. We need to set up a set of models that
test whether dN/dS along one branch is the same as another branch or not.
To do this, we first divide the
tree into two C sections, C0 and C1, and two A sections, A0 and A1, as
indicated above. We next set up
four hypotheses, H0 , H1, H2, and H3,
that make assumptions about the relative values of dN/dS among these four tree
components. For example, the null hypothesis, H0 , asserts that the
dN/dS ratios (omega values) along all branches are equal – i.e., no
change in evolutionary rates after duplication. In PAML, we use the control file parameter model=0 to test this
hypothesis that all rates are equal (see the control file below). Against this
hypothesis we test the claims that (H1) the ratio along C0 (along
the line after the presumed duplication) differs from the others; that (H2) the C lineage rate (C0 and C1) as a
whole is different from the A side of the tree (A0 and A1); and (H3) A1 and C0 are both
different from A0 and C1.
In order to do this analysis in PAML, we must supply a tree
structure file along with our usual control file and sequence file. Such PAML analyses require branch-specific
codon models to allow different branch groups to have different ωs,
leading to so-called “two-ratios” and “three-ratios” models. All those models require branches or
nodes in the tree to be labeled.
Branch labels are specified via the symbol “#” placed in a
parenthesis-notation format for the tree structure. The branch labels are consecutive integers starting from 0,
which is the default and does not have
to be specified. For example, the
following tree:
((Hsa_Human, Hla_gibbon)#1,((Cgu/Can_colobus,Pne_langur),
Mmu_rhesus),(Ssc_squirrelM, Cja_marmoset));
has a branch label #1 for fitting models of different ω
ratios for branches. The internal
branch ancestral to human and gibbon has the ratio ω1, while
all other branches (with the default label #0) have the background ratio ω0. This fits the model in table 1C for the
small data set of lysozyme genes in Yang (1998). (See the readme
file for PAML here.)
Yes, this is incredibly baroque and brain-dead. In fact, you really don’t have to
understand this notation to do the problem – the tree files are already
provided for you. An easy way to
visualize the parenthesis structure (if you are not used to Lisp), is to simply
input the tree description file into the program treeview,
which is on your CD-ROM. This is
how we produced the pictures for the different hypotheses below.
Here is the first part of the control file for the relevant codeml program:
seqfile = seqfile.txt * sequence data filename (replace w/ actual seq)
treefile = tree.txt * tree structure file name (replace
w/ tree file)
outfile = results.txt * main result file name
noisy = 9 * 0,1,2,3,9:
how much rubbish on the screen
verbose = 1 * 1:detailed
output
runmode = 0 * 0:user
defined tree (so you need to input a tree file)
seqtype = 1 * 1:codons
CodonFreq = 2 * 0:equal, 1:F1X4, 2:F3X4, 3:F61
model = 0 * 0:one omega
ratio for all branches
* 1:separate omega for each branch
* 2:user specified dN/dS ratios for branches
NSsites = 0 *
icode = 0 * 0:universal
code
fix_kappa = 0 * 1:kappa fixed, 0:kappa to be
estimated
kappa = 2 * initial or
fixed kappa
fix_omega = 0 * 1:omega fixed, 0:omega to be
estimated
omega = 0.2 * initial omega
------------
end of control file ------------------------------------
Start by specifying model=0, which sets all omega values the same on each branch, i.e., hypothesis H0. You must also input the tree file as specified in the control file. For the next 3 runs, you should set model=2, i.e., user specified omega values (dN/dS ratios) for specified branches in the phylogenetics tree. For these 3 runs you therefore provide a slightly different tree input file that says exactly where the dN/dS ratios are to be compared. These files are already in the lab directory for you to use:
Ldh codeml sequence file: http://web.mit.edu.ezproxyberklee.flo.org/6.891/www/lab/paml_ldh_seqfile.txt
Control
file: http://web.mit.edu.ezproxyberklee.flo.org/6.891/www/lab/paml_ldh_codeml.ctl.txt
Tree
H0 file: http://web.mit.edu.ezproxyberklee.flo.org/6.891/www/lab/paml_ldh_tree.H0.txt
Tree
H1 file: http://web.mit.edu.ezproxyberklee.flo.org/6.891/www/lab/paml_ldh_tree.H1.txt
Tree
H2 file: http://web.mit.edu.ezproxyberklee.flo.org/6.891/www/lab/paml_ldh_tree.H2.txt
Tree
H3 file: http://web.mit.edu.ezproxyberklee.flo.org/6.891/www/lab/paml_ldh_tree.H3.txt
Tree format for model H1:
(X02152Hom,U07178Sus,
(M22585rab,
((NM017025Rat,U13687Mus),
(((AF070995C,(X04752Mus,U07177Rat)),
(U95378Sus,U13680Hom))#1,(X53828OG1,U28410OG2)))))
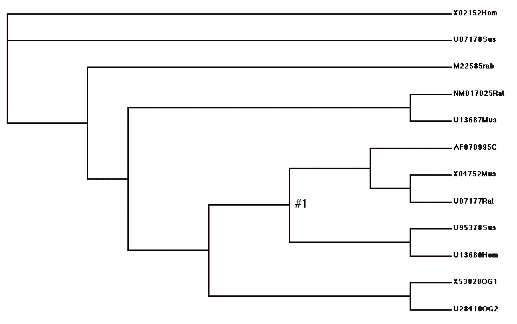
Picture of model H2. Note how this lumps all the “C” lineage together, and tests that omega value against the “A” lineage.
(X02152Hom,U07178Sus,
(M22585rab,
((NM017025Rat,U13687Mus),
(((AF070995C
#1,(X04752Mus #1,U07177Rat #1)#1)#1,
(U95378Sus #1,U13680Hom #1)#1)#1,
(X53828OG1,U28410OG2)))))

Your job is to run these analyses to fill in the following table where the ‘blank’ values are and confirm (or not), the questions that were given at the beginning of this section.

![]()
![]()
![]()
![]()

![]()
Part 4:
Application to Yeast (tbd)